
|
TRAD
TRAD (Tip-Rooting and
Ancestor-Dating)

| Name: | TRAD |
| Main function: | As suggested by the title, used for rapidly
evolving viral strains collected over different times, without a good outgroup |
| Requirement: | (If working with a tree with >60,000 leaves): a
fast desktop computer with at least 8GB RAM |
| Keywords: | phylogenetics, dating, rooting, virus |
| Version: | 1.0.0.0 |
| Publisher: | XiaLab |
| Support: | Dr. Xuhua Xia at xxia@uottawa.ca |
| Citation: | - Xia, X. 2025 On rooting and dating viral trees with a changing evolutionary rate following host-switching. Genome Biology and Evolution
- Xia, X. 2025. TRAD: Tip-Rooting and Ancestor-Dating, version 1.2.0. XiaLab
|
 Input for TRAD:
Input for TRAD:
- The ncov-2019.ml.tree by Andrew Rambaut. This maximum likelihood tree is based on 189 early SARS-CoV-2 genomes. The original download site is here
- An alternative tree TREE188_PhyML_NoNodeLabel.dnd that is similar to the above, but reconstructed after removing one poor-quality genome. The remaining 188 genomes were realigned with MAFFT and a maximum likelihood tree was reconstructed with PhyML. You may also download the sequence alignment here in FASTA format
- Any reliable phylogenetic tree with the collection date appeneded to OTU names (e.g., USA/IL1/2020|EPI_ISL_404253|USA|Illinois|Chicago|2020-01-21). Alternative date formats are acceptable.
 TRAD for Windows PC: TRAD for Windows PC:
- If your Windows PC already has .NET 8 installed, download and run TRADSetup.msi,
otherwise download and run Setup.exe. The default installation folder is C:\Program Files\XiaLab\C:\Program Files\XiaLab\TRADSetup. Click TRAD.exe to start.
- Once TRAD starts, click "Phylogenetics | TRAD", input a tree, e.g., ncov-2019.ml.tree, set the options, and click "OK".

 TRAD for Macintosh TRAD for Macintosh
- Download Whisky, unzip it and copy Whysky to the Application folder.
- (Let me know if the Whisky link above becomes unavailable)
- Download TRAD.msi
- Double-click Whisky and follow the concise instructions to run the downloaded TRAD.msi file.
- Whysky includes a "winetricks" button. Click it and click "Dlls" tab. Select "dotnet48" (which means .NET Framework 4.8) and click "Run". It could take 10 minutes to finish installation. A Terminal window will display the program. Wait until the Terminal window returns to the Linux command prompt.
- You can now double-click "TRAD" (which has a green arrow above) to run TRAD. You may need to wait for 10 seconds the first time you do it. During this ~10 seconds, there seems no response, but TRAD will eventually start.
- Once TRAD starts, click "Phylogenetics | TRAD", input a tree, e.g., ncov-2019.ml.tree, set the options, and click "OK".
Here are some screenshots of running TRAD on an M2 MAC (M1, M2, M3 and M4 MACs work the same way).
The first dialog for inputing a tree for TRAD:
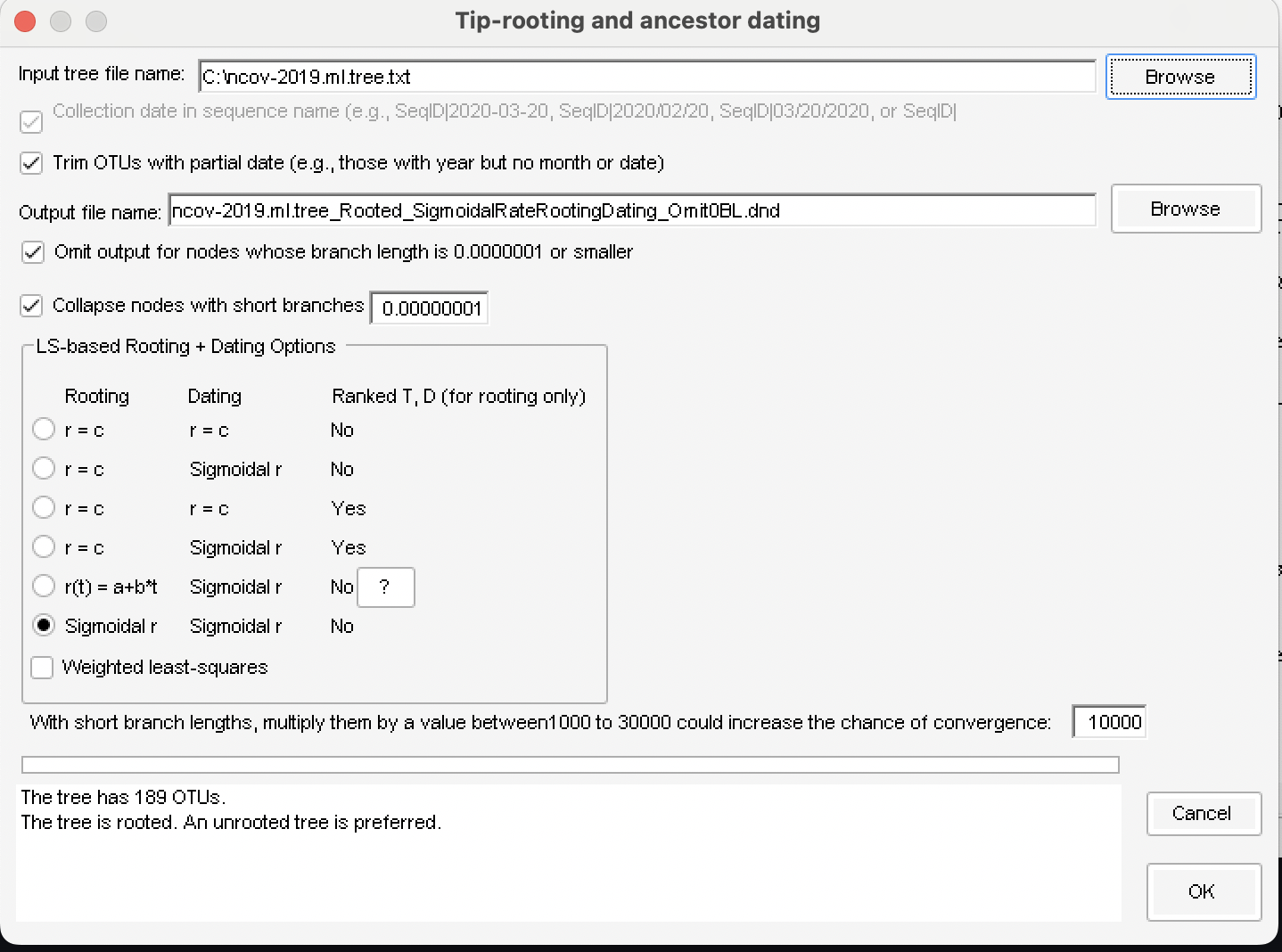
The observed and fitted relationship between D (root-to-tip distance) and T (Collection time)
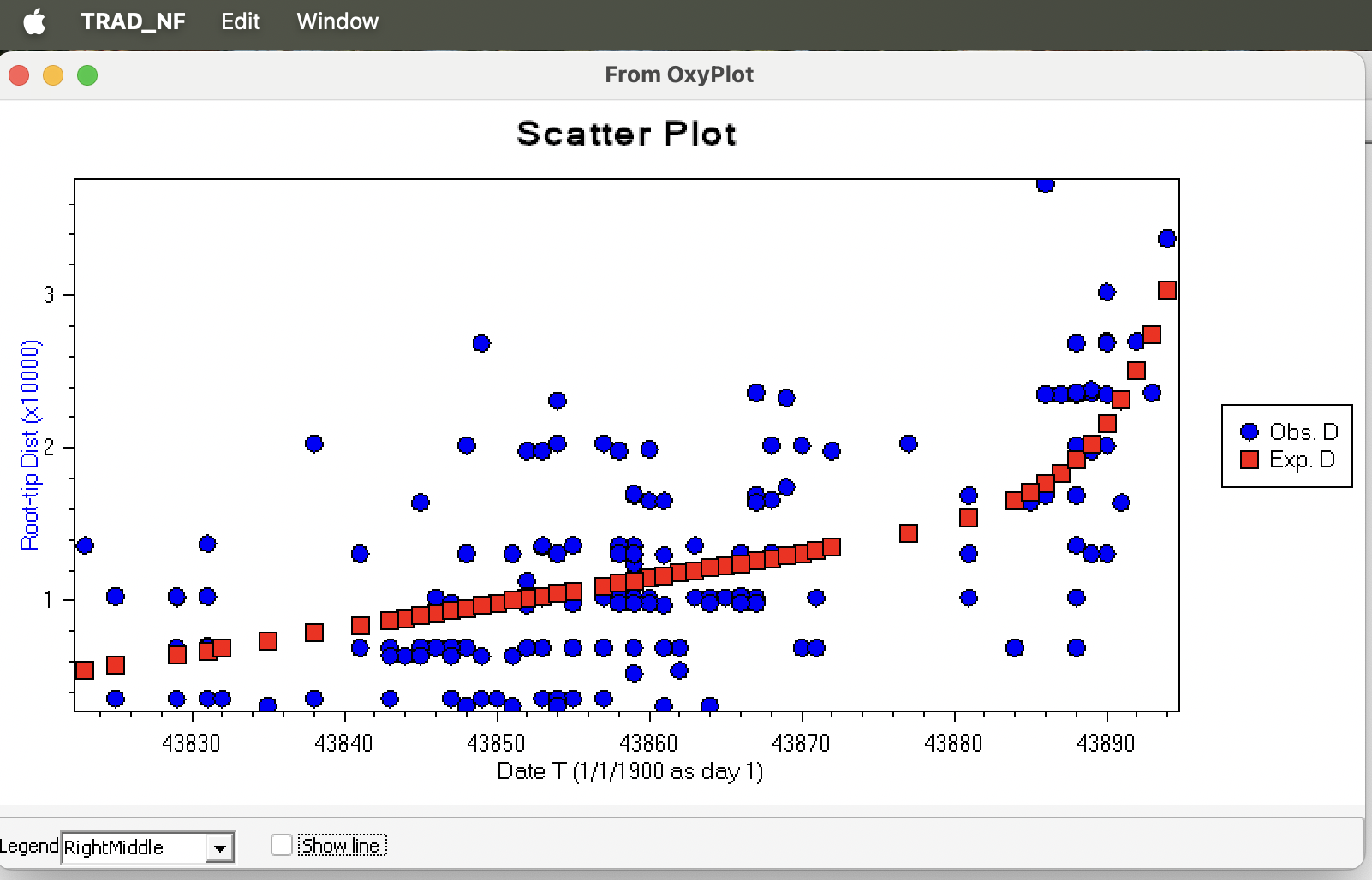
The predicted r (evolutionary rate) and T (Collection time):
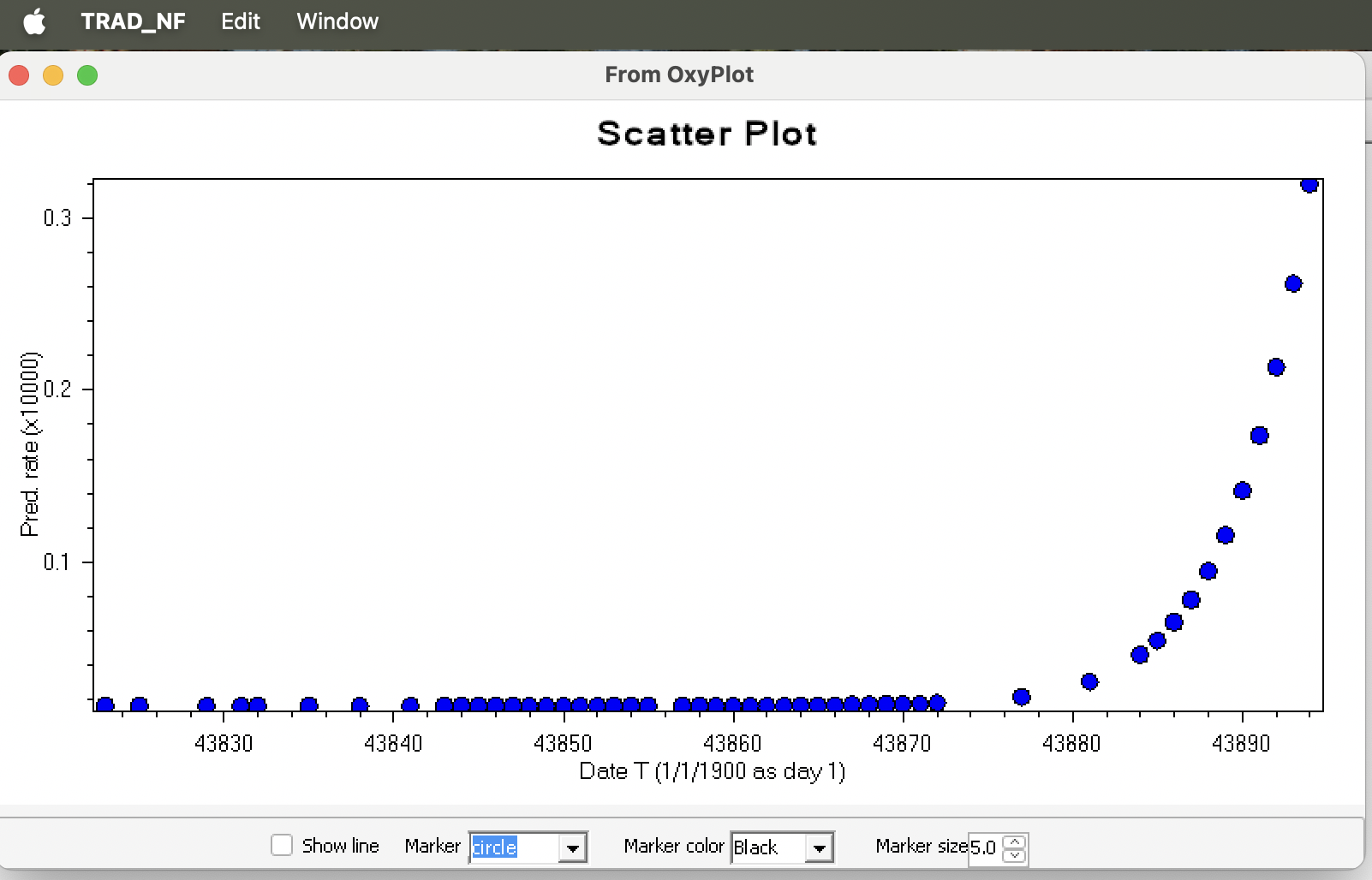
The rooted and dated tree:

 For 64-bit Linux computers: For 64-bit Linux computers:
- Click the App Store or Package Manager, and choose wine to install.
- (alternatively, issue the command sudo apt install --install-recommends winehq-stable)
- At the end of wine installation, you may be prompted to instlal Mono. Acept to install
- Download TRAD.msi
- wine msiexec /i TRADSetup.msi
- TRAD is now ready to run.
- Here are more information on msiexec

Usage after installation:
- Click 'Phylogenetics | TRAD
- Input: a phylogenetic tree in Newick format, with collection time at the end of the sequence name. A test file Test_Fig1.dnd is provided.
- Output: evolutionary rate, the time of origin of the most recent common ancestor, the most likely rooting point
Warning:
- GenBank contains fake SARS-CoV-2 genomes. See my paper for details. You should not include such genomes in your analysis.
- You should include only viral sequences isolated from natural hosts, not from specially treated experimental animals.
- Do not include genomes from viruses that have been preserved for a long time (e.g., by deep-freezing) and then thawed and used in experiments. Such viruses have been frozen in evolution. Their genomes will mislead you to a very slow evolutionary rate and erroneous dating of the common ancestor.
|